
A techbio company not using AI today would be like a tech company not using the cloud.
Biology is getting digitized. For the past decade we’ve developed richer, bigger datasets – from genomics, proteomics, transcriptomics, to images of young and old cells. AI was needed to make sense of it all. The rise of AI in bio was inevitable.
Right now, generative AI is the new hotness across industries. There’s a good reason: this technology is amazing. It feels like magic. But while AI is the new buzz for in more fields, it’s already been around for quite a while in bio.
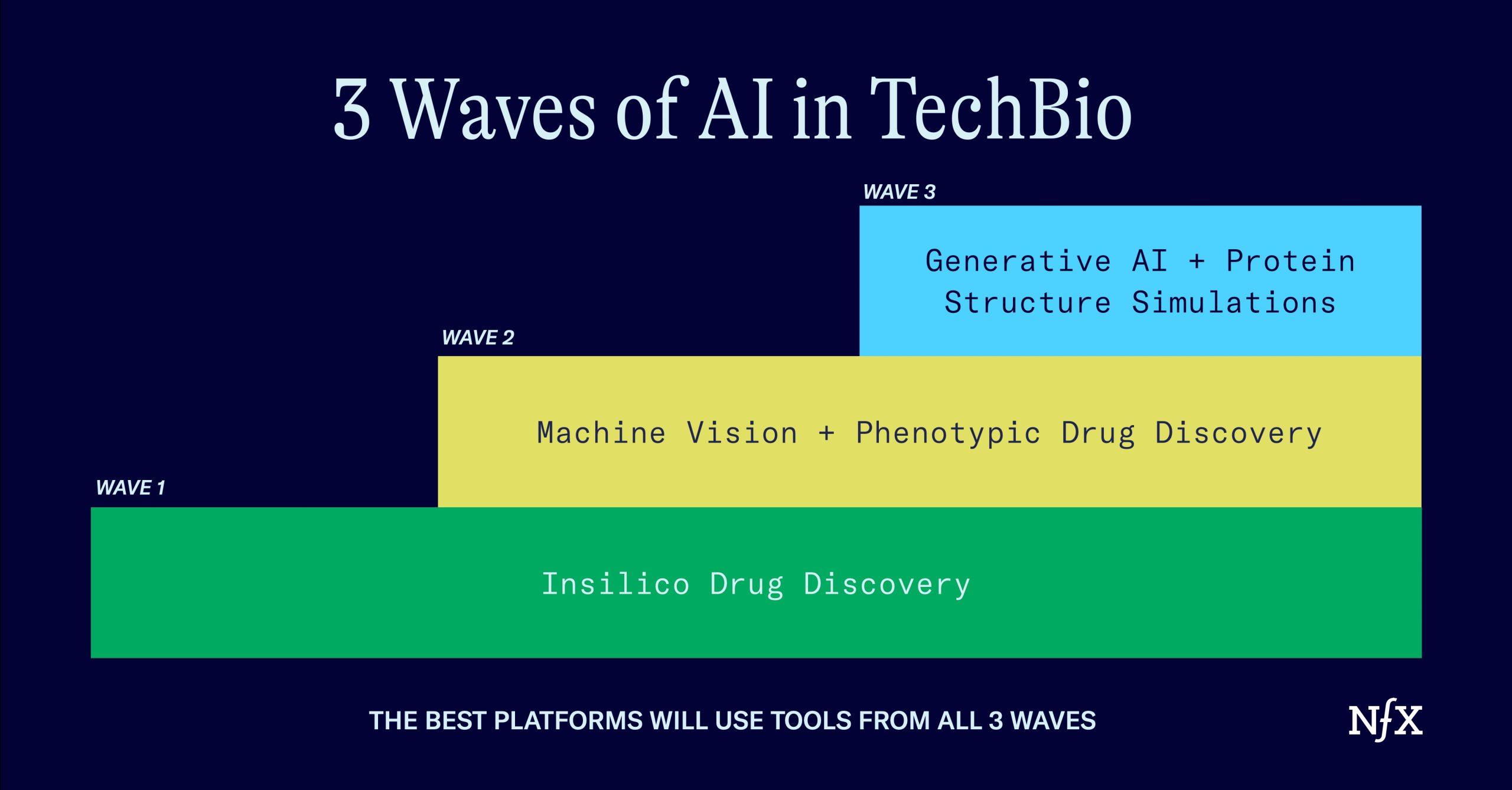

We’ve seen at least three waves of AI in bio:
- In-Silico Drug Discovery
- Unlocking Phenotypic Drug Discovery
- Generative AI + protein structure simulation
Once you understand these three waves, you’ll be able to see how they all fit together to create the next massive wave of techbio companies (and what traps founders and investors need to avoid along the way).
Wave 1: In-Silico Drug Discovery
An early wave of AI in techbio was about scaling up the compound screening process.
In 2012, Atomwise was one of the first to get into this game. Atomwise trained AtomNet, their own Convolutional Neural Network, on huge amounts of biological data on molecular structure and function.
Convolutional Neural Networks were the hot technology at the time. But the real insight was in the use case for these networks. In the past, you had to actually solve the structure of proteins, examine how small molecules bind (or don’t) to certain targets, and run huge screens manually.
Instead, this early wave AI in bio allowed for the development of in-silico screening using experimentally derived molecular structures. It opened up the ability to computationally test millions of compounds on any specific experimentally solved protein target.
Atomwise gained a huge first mover advantage by getting into this game early. They got funding from tech sources like Y Combinator. They got the big pharma deals. By the time many other companies were aware of this potential, this first wave had already swept through.
This in-silico approach was a huge breakthrough. But it’s not perfect. This process added lots of drug candidates to the top of the clinical trial funnel, but it did little to help advance those candidates through the clinical trials which is where most of the cost is anyway.
Today, this space is extremely crowded. To stand out, the next wave of companies needed to build on top of this idea, or unlock a new way of getting better drug candidates into clinics even faster.
Wave 2: Machine Vision Unlocks Phenotypic Drug Discovery
Around 2016, the second wave of AI in bio unlocked phenotypic drug discovery using machine learning and machine vision. Computers became observational tools that could tell humans what compounds actually work to reverse disease, and which don’t. It’s a far more efficient form of experimentation.
This wave was still based on the underlying technology of Convolutional Neural Network. These networks are all about teaching a computer to learn the “essence” of things. When we see a dog or a cat, our brain intakes data from the world, processes it, and generates an output. We recognize that object as a dog or a cat. But all a computer sees are pixels.
Over time, though, if you show a computer enough pictures of a dog or a cat, and tell it what’s what, it starts to learn the rules. Certain pixel values organized in certain ways = a dog. Other pixel values organized in other ways = a cat. The computer learns what value equals what output. The essence of “dogness” or “catness.”
We have trained AI on models of disease in a similar way. Computers began to recognize signs of aging or unhealthy cells. It allowed us to see which compounds might revert those sick or aging cells back to a younger, healthier state en masse.
An example of a company in this space is Spring Discovery. Spring Discovery trained an algorithm on images of millions of old and young cells to find the key pixels that distinguish old from young.
Combined with proteomics data, Spring Discovery is allowing AI to find patterns in cell phenotypes that humans can’t. They describe it similar to Star Trek’s tricorder – the AI senses patterns that we would never see.
This approach has allowed Spring Discovery to tackle increasingly complex forms of disease – like aging, which is dependent on variety of different mechanisms in the body. Spring Discovery is approaching aging by focusing on cell phenotypes, rather than by individual mechanisms.
Another example of a company tapping into the power of machine vision and images as data is Recursion Pharmaceuticals. Recursion’s machine vision algorithms are trained to distinguish healthy and diseased cells in high levels of detail. Then, scientists can expose thousands of diseased cells to potential drug candidates, take snapshots of the results, and identify which compounds reverted cells from the diseased, to healthy states.
This second wave of machine vision has allowed us to ask questions more broadly. You don’t have to dig deeply into the mechanisms of why certain drugs work and don’t work. You can focus on what works right away, and move ahead fast. And, computers can spot patterns humans could never see on their own.
There’s a lot of potential in this approach. But now we’re seeing that you can do much more when you add this wave together with what’s coming next…
The third wave is the next logical step: moving from understanding and analysis to prediction and generation.
Wave 3: Generative AI + Protein Structure SSimulations
We’re now in the third wave of AI in techbio: AI now sees patterns and generates something entirely new.
Three big things came together to get us here.
First, DeepMind’s Alpha Fold achieved the previously unimaginable, and predicted the structure of nearly every protein cataloged by science. This advancement of being able to simulate protein structure was also achieved by several other players around the same time: Meta, and David Baker’s lab at the University of Washington. It was a watershed moment in the field.
Then, Large Language Models pushed the field forward even more. LLMs like ChatGPT are trained on all of human writing, and can predict the next word in a sentence. LLMs in biology were trained on all the different open reading frames of DNA that have evolved over 4 billion years. Similarly, they became capable of predicting the next amino acid in a protein’s sequence with a high degree of accuracy – the same way ChatGPT predicts the next word in a sentence.
Finally, David Baker’s lab at UW has repeatedly proven that de novo protein design software is a viable approach.
Taken together, these advances were circling around a big idea: that we could one day design proteins from scratch, rather than just discover and tweak them.
The combination of these factors has paved the way for our new ability to generate new things. We have models that “understand” the core of protein-ness, from the biological language that makes them up, to the structures that facilitate function. And we are developing the tools to design them from scratch.
NFX-portfolio company Zip Therapeutics has developed a machine learning engine that can take a large protein amino acid sequence, understand the most valuable parts for function, and condense that protein into a smaller, but fully functional form.
The result: a new protein that has never existed before in nature, but is still effective. And, critically, small enough to fit into an adenovirus vector (solving a delivery bottleneck for certain monogenic diseases). That’s the key part. Zip has used this AI tool to solve a uniquely challenging problem.
Another example is Nabla Bio. Nabla Bio has used LLMs to design, rather than discover, new proteins. Nabla can express millions of potential antibodies in a single tube, measure for specific therapeutic properties, and reverse engineer the sequences that lead to those properties. That data loop then allows for new, optimized antibody design.
This is an exciting time for AI in many fields. But for those of us working in techbio, generative AI in bio is really just another (powerful) tool that is going to be used by the majority of techbio companies. If you want to stand out, you have to think about unique and defensible ways to deploy this tool, and all those that came before it.
Building a Company in Wave 3
Today, no one would market themselves as a company using Excel spreadsheets. It will be the same with generative AI in bio. As with the previous first two waves in bio, these new developments rapidly become the new normal tools that everyone learns how to use.
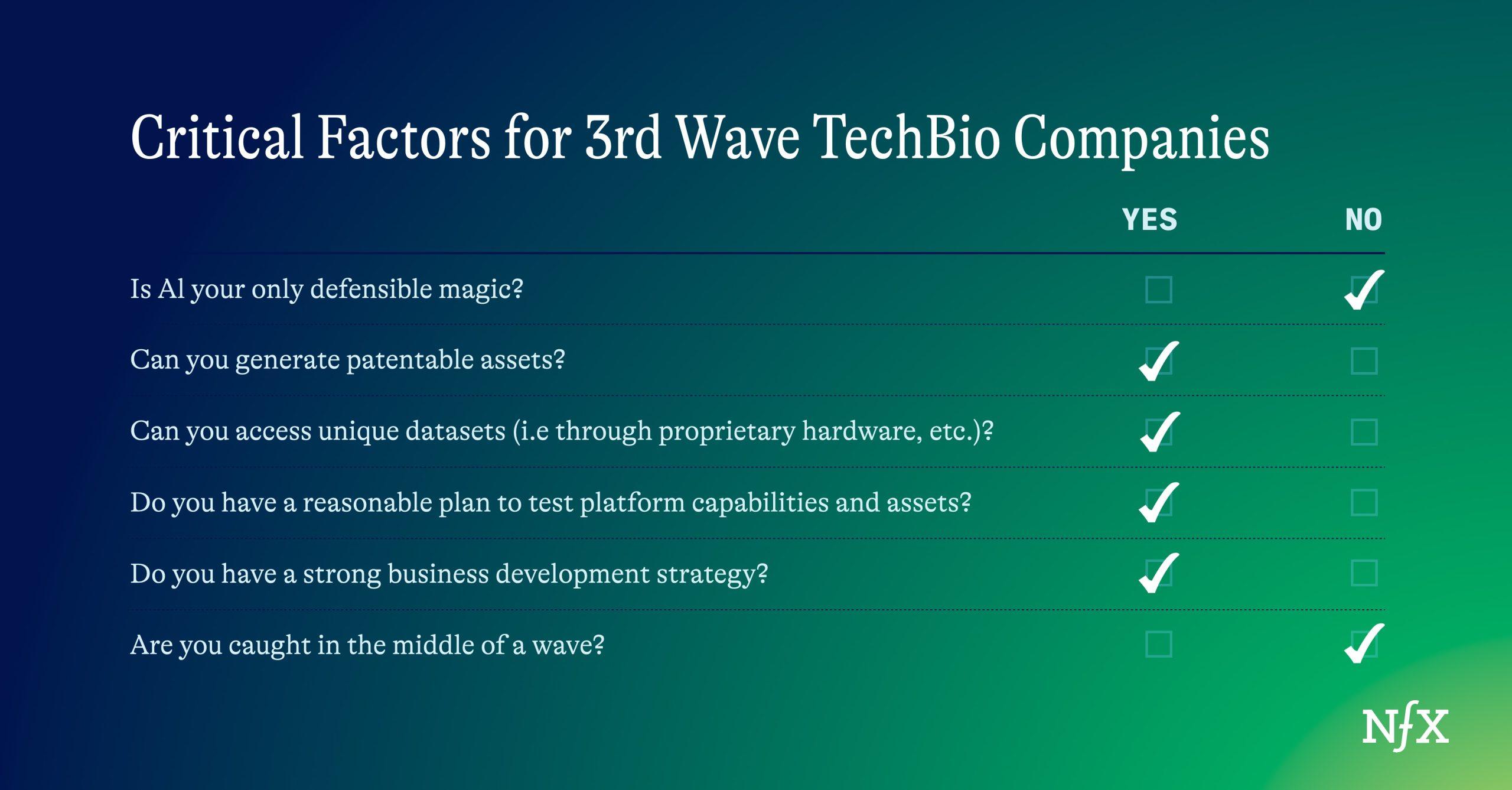

To stand out, you need to consider five things:
1. AI is magical, but it is not your defensible magic
We look at three things when we evaluate companies:
Are you the right team?
Is your problem big enough?
And do you have defensible magic?
Many companies believe that their AI is their defensible magic right now, but that’s not going to last. This pattern will unfold across many other industries, as we’ve described before. Using an AI tool right now feels like magic. But these models are easy to copy, as long as you have insight into the inputs and outputs.
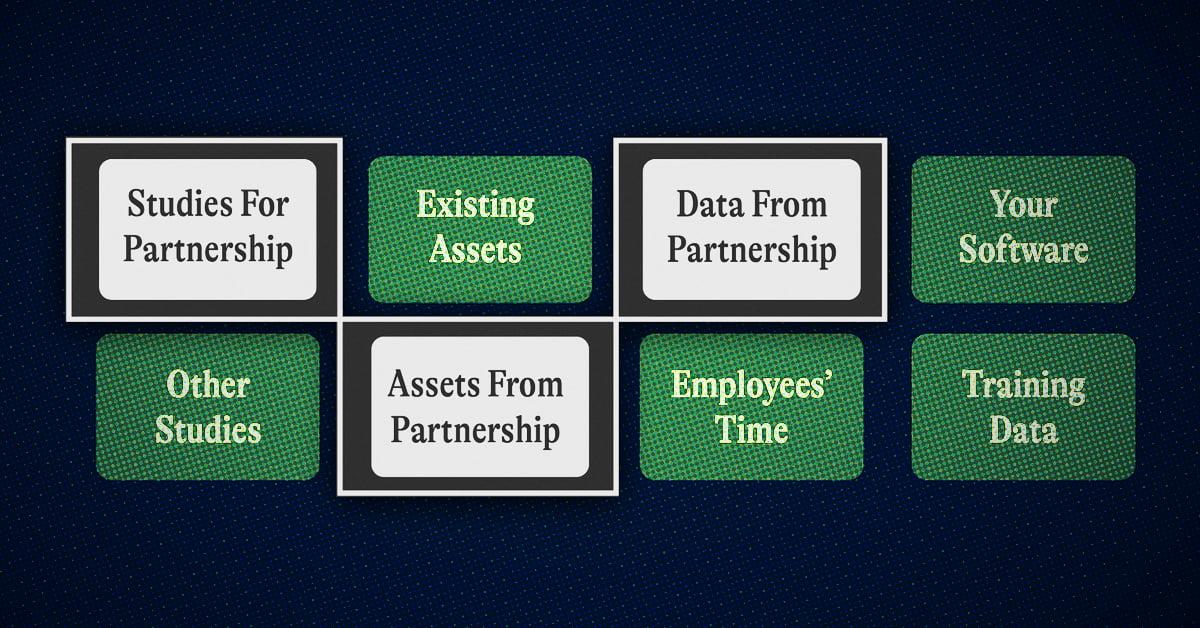
There will be some exceptions. For example, if you can access data that is extremely hard to find or have hardware that allows you to capture a novel form of data you might be able to stay afloat for a while. NFX portfolio company Pumpkinseed, for example, is developing a new silicon chip that can capture proteomics data that has been previously impossible to collect.r
But the truth is: AI alone isn’t your secret sauce. The AI must allow you to do something that no one else can do. This leads to our second piece of advice:
2. Generate something patentable
If your assets work, they will win. No one cares if you generated it from your grandmother’s cookbook or the Oracle of Delphi or a new generative AI model. If you can patent it, it doesn’t matter where it came from.
That’s how your partners will measure the value of your platform, especially in this current funding environment.
It’s how you should be thinking too.
3. Can you generate things that work? Can you prove it?
Lots of people will be able to generate novel compounds very soon. But biology isn’t usually plug and play – for unknown reasons some of these compounds just aren’t going to work in practice.
Make sure you’ve thought through how your newly generated projects work in a larger context. Can you actually deliver them to the organs they need to get to? Can you test them? Can you synthesize them? Can you rule out the non-viable candidates fast? How long is your iteration cycle?
These are the questions you’ll need to answer step by step. You’ll need a strategy for building a clinical results warchest that answers each of these questions in turn. Find our advice for that here.
4. Don’t get caught in the middle
Being first into a new wave is good – the first wave of companies using AI for drug discovery got lots of deals with pharma.
It’s also okay to be last, in some cases, because you can piece together many waves of new technology in its best form. NFX-backed Pepper Bio waited until the fields of proteomics, transcriptomics, phosphoproteomics, and genomics had advanced and built a platform to work across all the layers.
Being in the middle rarely works.
The second generation of AI for drug discovery found it hard to break out. There was too much noise. The same will be true for generative AI.
5. Win on the Business Side
The best companies really understand the business. You need to have amazing technology. But you also need to understand how to sell yourself: to investors, to pharma, to patient groups etc.
You don’t even have to know how to market your whole platform. You just need to know how to sell one or two things that you can do better than anyone else, and that people are willing to pay for.
When you’re looking at building partnerships for your platform ask yourself: Who wants to buy my service? How can I sell to them? More on both of those questions here.
Understanding Tech + Bio is Even More Necessary
Techbio companies are already deeply familiar with AI. But what AI allows us to do in the field is changing fast.
Because of this, we’re going to need more teams that consist of both tech people and scientists. There’s never been a better time for these two kinds of people to build a company.
We’ve been on both sides. Come talk to us.
As Founders ourselves, we respect your time. That’s why we built BriefLink, a new software tool that minimizes the upfront time of getting the VC meeting. Simply tell us about your company in 9 easy questions, and you’ll hear from us if it’s a fit.